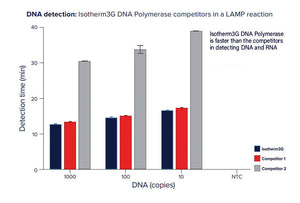
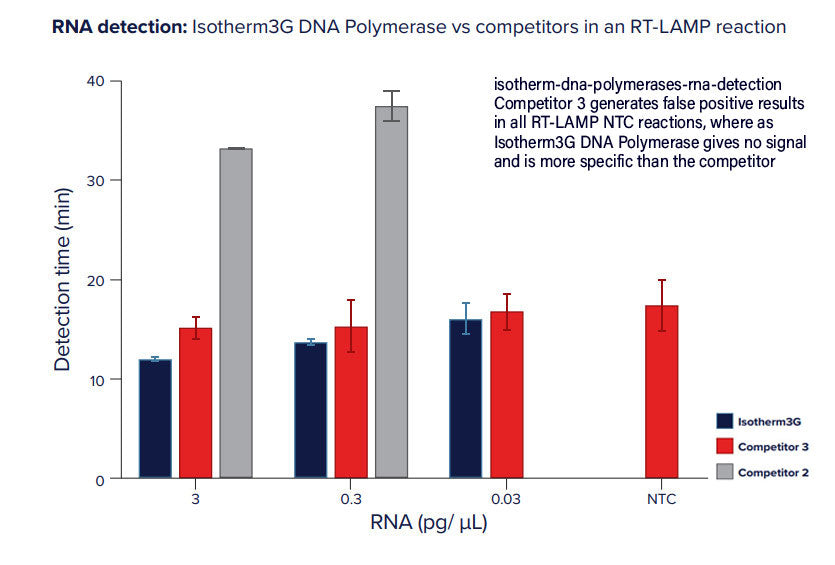
RevTaq RT-PCR DNA polymerase
-
RevTaq RT-PCR DNA polymerase is an engineered, extremely thermostable reverse transcriptase and combined DNA polymerase, obtained through directed, artificial evolution.
RevTaq RT-PCR DNA polymerase can be used similarly to Tth DNA polymerase also for reverse transcription, but does not require the addition of Mn2+ for RT-PCR, which simplifies assay setup and possible sample processing.
- half-life at 95°C of >40 min.
- fast start function due to a hotstart aptamer formulation which prevents unspecific amplification at lower temperatures (<57°C)
- facilitates “zero-step” RT-PCRs directly from RNA templates (without an isothermal reverse transcription step), as reverse transcription takes place simultaneously with DNA amplification during the cycled PCR elongation step.
- allows reverse transcription reactions at high temperatures, thus minimizing the problems encountered with strong secondary structures in RNA that melt at elevated temperatures.
- RevTaq RT-PCR DNA polymerase is the pure, reverse transcription active enzyme ingredient of our Volcano3G® RT-PCR Master Mixes
Please note: Due to the thermostable nature of the enzyme, it is recommended to design very high melting primers and probes (>65°C). See more details in FAQ.
For research use and further manufacturing.
In case you are aiming to use our RUO products as components or for your development of e.g. an IVD medical device, please contact us.
-
- RNA into DNA and PCR in one step
- Rapid detection and identification of RNA & DNA target
- Reverse transcription qPCRs (RT-qPCRs)
- Direct from cell RT-PCRs
- qPCRs
-
RevTaq RT-PCR DNA polymerase is tested for a successful, reproducible RT-qPCR performance. Enzyme concentration is determined by protein-specific staining. The activity of the enzyme is monitored and adjusted to a specific DNA polymerase activity using an artificial DNA template and DNA primer.
Additionally, a 151 bp fragment (HPRT1 mRNA) is amplified from an in-vitro transcribed RNA strand by a 0-step real-time RT-PCR protocol using hydrolysis probes and the linearity of amplification over a specified serial dilution is demonstrated. No human RNA or nuclease contaminations is detected.
This product is for research use only. Please get in touch with us if you need it for diagnostic or commercial purposes.
-
How do I have to store the polymerase?
- Keep at -20°C
What special instructions must be observed?
- Due to the aptamer-based hotstart-formulation, RevTaq RT-PCR DNA polymerase will yield better results with annealing and extension temperatures above 57°C.- Due to the thermostable nature of the enzyme, it is recommended to design very high melting primers and probes (>65°C).
- It is recommended to optimize the temperatures of the annealing/extension steps by applying a temperature gradient during establishments.
- Higher temperatures lead to higher specificity of the PCR. The RT-cycles are usually run at higher temperatures than the PCR cycles. This is due to the fact that DNA-primer: RNA-template hybrids have generally higher melting points than DNA-primer: cDNA template duplexes.
- Depending on the employed assay, the reverse transcription steps may be omitted (zero-step RT-PCR).
- RevTaq RT-PCR DNA polymerase is engineered and optimized for an amplicon size between 60- 300 bp.
-
Establishing RT-PCR assays with RevTaq RT-PCR DNA polymerase:
1. Make sure that your primers have melting points higher than 65°C
2. Optimize PCR-steps: Prepare one big PCR reaction with cDNA and aliquot it into the plate in multiple wells. Apply a two-step PCR protocol (for example 95°C 10 sec. and a temperature gradient between 57-67°C (1min, 40 cycles).
> From this experiment you can select the best temperature/well which shows the earliest Ct-value.
3. Optimize cDNA generation/reverse transcription steps: Then, take an RNA sample with a relatively high amount of target RNA (~107-105 copies/rxn) and run an RT-PCR with a protocol such as this: 10 cycles of 95°C 3sec and a temperature gradient between 60-75 °C for 1 min) and followed by PCR cycles with the best PCR temperature as selected in step 2.
> From this experiment you can select the best temperature/well with the earliest Ct-value.
Further optimizations:
These two steps above combined should already result in a very decent RT-PCR protocol.
Finetuning is possible by shortening the extension-times during PCR phase or further optimizations of the denaturation temperatures (via temperature gradient RT-PCRs e.g. between 85-100°C).
Related Items
Our quality management system is certified according to ISO 13485.
myPOLS Biotec is part of Medix Biochemica Group.
Phone: +49 (0)7531 122 965 00
Fax: +49 (0)7531 122 965 02
You find us at Byk-Gulden-Straße 2
78467 Konstanz, Germany
Contact us.
Sign up to get the latest on sales, new releases and more...
Disclaimer & Data Privacy Statement
© 2024 myPOLS Biotec.